Senescence
Senescence is the permanent halt of proliferation associated with cellular stresses including DNA damage, damaged or short telomeres and oxidative stress. Senescence dates back to the Hayflick limit and, while there are no definitive markers, cells typically become large and flat and can be identified by the presence of SA-β-GAL, SAHF and tumour suppression networks (i.e. p53 / p16).Senescence is thought to be a tumour suppression mechanism, through blocking clonal proliferation of cancer cells, and a promoter of ageing via an inflammaging phenotype. Cancer cells must somehow bypass senescence in order to continue proliferating.
Methylation
We were interested in studying genome-wide DNA methylation patterns within senescence. DNA methylation is the addition of a Methyl group to specific DNA bases. In mammals this is restricted to the cytosine nucleotide (C) within DNA. Methylation is largely localised at CpG's (a cytosine nucleotide followed by a guamine nucleotide) within the genome.In order to study DNA methylation we use whole genome sequencing combined with bisulphite treatment. Bisulphite treatment of DNA converts unmethylated cytosines into uracil - when we apply a process known as polymerase chain reaction (PCR) to the DNA before sequencing this is then further converted into thymine. Methylated cytosines are unaffected. Therefore, when sequencing a methylated cytosine will be read as a cytosine while an unmethylated cytosine will be read as a thymine. As such when we sequence the DNA we can then deduce, via comparison with the human reference genome, which cytosine nucleotides were originally methylated and which were unmethylated.
Loss of Methylation at Late Replicating Regions
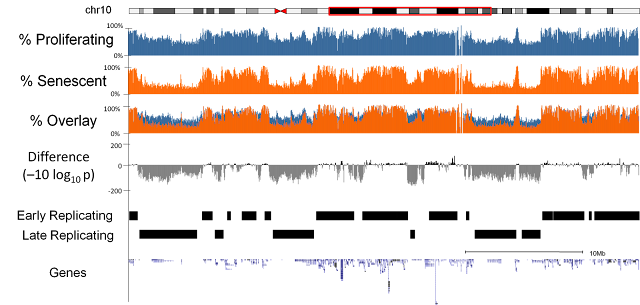
We sequenced three replicates of proliferating and replicative senescent cells (IMR90 cell culture). Our first observation was that there was a general loss of methylation (hypo-methylation) within the cells and that this loss was focused at regions of the genome which were only partially methylated (as opposed to completely unmethylated or fully methylated). Further investigation highlighted a relationship with the regions losing methylation and late replicating regions of the genome. i.e. regions of the genome where DNA is replicated later on during the cell cycle[1].Focal Hypermethylation
We observed that methylation does not globally regulate gene expression changes but we did detect a subset of genes with very similar patterns in terms of methylation changes and which were coregulated at the expression level. In these genes we observed focal increase in methylation (hyper-methylation) at the regions adjacent to islands of high CpG content and that these genes were frequently associated with cell cycle functions.Relationship to Cancer
Surprisingly, considering the role of senescence as a tumour suppression mechanism, we discovered that the methylation changes in senescence were very similar to the methylation changes that had previously been described in cancers[2,3,4].Origins
We discovered that the origin of the loss of methylation appeared to be due to mislocalisation of the DNMT1 methyltransferase which is responsible for propagating the original methylation pattern of DNA at replication. Specifically we noticed that foci of the DNMT1 protein are lost in old cycling cells and that these foci had previously been linked to late replicating regions[5].Bypassing Senescence
Finally, we bypassed senescence via the introduction of SV40 T antigen into the cells in order to deactivate tumour suppression networks. This allows the cells to continue proliferating, for a period of time, beyond the usual senescence time limit. When we did this we made the same measurements of DNA methylation as before and found that the methylation changes that occur on the path to senescence are retained in the bypass cells.Conclusion
We therefore conclude that the failure of DNMT1 to methylate late replicating regions in near senescent cells may be the origin of the same methylation patterns found in cancer cells. Furthermore, given the similarities, it may be possible that these changes may lead cells to be more susceptible to transformation into cancer cells.The full paper is available online from Nature Cell Biology, is discussed by a CRUK press release and the raw data is available from GEO GSE48580.
References
[1] R. S. Hansen et al., Sequencing newly replicated DNA reveals widespread plasticity in human replication timing, PNAS, 2010.
[2] K. D. Hansen et al., Increased methylation variation in epigenetic domains across cancer types, Nature Genetics, 2011.
[3] B. P. Berman et al., Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina–associated domains, Nature Genetics, 2012.
[4] G. C. Hon et al., Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Research, 2012.
[5] L. Schermelleh et al., Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res, 2007.







Good article. Thank you.
ReplyDelete